What technique was probably used to measure the f-state
MATCHING
Match the words with the definition or property. Only one correct answer
protein stability a. major driving force for polypeptide compaction
junction b. at this, the amount of folded and unfolded states are equal
turn c. helix geometry that cannot be found in RNA
B-form d. Always unfavorable for folding
base step e. connects two elements of secondary structure using 1-2 residues
indirect readout f. sequence-specific contacts between amino acids and bases
motif g. bind to palindromic DNA sequences (inverted repeats)
tertiary interactions h. 3-D classification of polypeptide or nucleic acid
counterion condensation i. Binding specificity that arises from the sequence-dependence of deformations away from B-form
fold j. 3-D contacts between residues that are far apart in sequence
major groove k. result of London dispersion forces between planar bases
melting temperature l. energetics controlled by base-stacking interactions
direct readout m. compact regular structures with repeating backbone torsion angles and maximal backbone hydrogen bonds
?Cp for folding n. proportional to surface area difference in F and U states
Hydrophobic effect o. describes a general advantage for binding polycations to DNA due to entropy
DNA bends p. two consecutive basepairs
homodimers q. a short sequence associated with a fold or function
stacking interactions r. Difference in free energy between folded and unfolded states
chain entropy s. where three or more helical nucleic acid segments are joined
secondary structure t. provides protein access to the bases in DNA helices
Folding Concepts In order to analyze energy differences between U and F states, it is critical to know the differences in their structural properties. Using understandable sentences or phrases (one or two dangling key words will be given NO CREDIT), DESCRIBE TWO UNIQUE STRUCTURAL properties that are relevant to their energy differences.
A. UNFOLDED PROTEINS
B. FOLDED PROTEINS
C. There are four thermodynamic functions (?Hprotein, ?Hwater, -T?Sprotein, and -T?Swater) that contribute to ?Gf in water. Provide a BRIEF molecular/atomic explanation for each:
5) Which one(s) is (are) ALWAYS favorable for protein folding at temperatures -50°C to 100°C?
6) Which one(s) is (are) ALWAYS unfavorable ?
7) Which one(s) is (are) responsible for unfolding at low temperatures? 8) EXPLAIN BRIEFLY WHY a protein unfolds at high temperatures?
Stability measurements and calculations
Below are the CD melting curves for a protein DUD1 and a mutant, dud1A, which contains a Cys54?Ala mutation. The heat capacity change for DUD1 folding is -1.5 kcal/mol-K, and assume it is the same for dud1A.
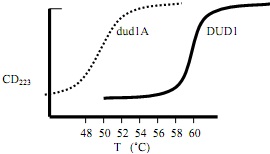
1) What is the Tm of DUD1 (nearest °K) ?
2) What is the Tm of dud1A (nearest °K) ?
3) Which protein is more stable?
4) For DUD1, the ?Hf at its Tm is -100 kcal/mol, but for dud1A, the ?Hf is -83 kcal/mol at its Tm. What is the change in ?Hf (??Hf) incurred by the mutation at the DUD1 Tm?
5) Is this change favorable, unfavorable or neutral?
6) At the DUD1 Tm, what is the value for ??Sf (not -T??Sf) incurred by the mutation?
7) Is this change favorable, unfavorable or neutral?
8) Based on your answers to 4-7, provide a reasonable explanation for WHY DUD1 and dud1A have different stabilities (what is Cys54 doing?).
Electrostatic Contributions to Stability
In a protein SGMD, there are two lysine residues, Lys25 and Lys11. The pKa of a Lys residue in a short peptide is 10. In the native protein, Lys25 has a pKa of 12, and Lys11 has a pKa of 10. Based on this data, answer the following questions
(SHOW WORK FOR CREDIT):
1) Estimate the unfolded (U) state pKa for each: Lys25 Lys 11 .
2) What technique was probably used to measure the F-state pKa?
3) Calculate/Estimate the contribution of the ionization of Lys25 to the folding free energy, in kcal/mol Hint: thinking about linkage makes this easy to do! Make sure that the sign correctly indicates favorability or unfavorability.
4) Calculate/Estimate the contribution of the ionization of Lys11 to the folding free energy, in kcal/mol
5) Briefly, describe the physical environment of Lys25
6) Briefly, describe the physical environment of Lys11
7) A glutamate side-chain carboxylate accepts a hydrogen bond one of these lysines. Which Lys?
8) This glutamate most likely has a pKa of 2.0 4.0 6.0
9) If you mutated this Glu to Gln, what is the effect on stability ?
10) In the Gln mutant, what would you expect to happen to the pKa of the lysine you chose in answer 5 ? ?
Mutations and Stability
For the two substitutions made at various locations in the folded state of a protein
a) on the graphs below indicate the relative changes in free energies of the folded (F) and unfolded state (U), using arrows (see example “Ex.”)
b) In the spaces below, for each state, U and F, BREIFLY EXPLAIN WHY the changes occurred (1-2 sentences, again, dangling phrases get NO CREDIT)
c) predict the overall change in stability, ?Gf (favorable/unfavorable)
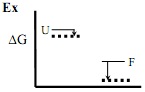
1). Surface Arg->Met a)
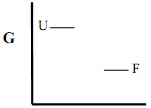
F why?
U why?
c) Overall ?Gf
why?
2) Buried Ile->Ser

b)
F why?
U why?
c) Overall ?Gf
why? _
3) Buried Val->Trp
In addition to changes in free energy (G), first illustrate and give reasons for the expected changes in enthalpy (H) and entropy (-TS) for U and F states and the overall changes (favorable/unfavorable), that generally accompany this type of mutation.
Enthalpy (Note T>20°C)
a)
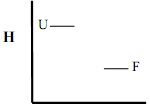
b)
F why?
U why?
c) Overall ?Hf
Entropy
b) F why? U why?
c) Overall -T?Sf
Free Energy
a) G

b) F why? U why?
c) Overall ?Gf
Sequence-structure-function relationships
Below are the sequences of DNA-binding domains for two different closely-related classes of the BDUN site-specific DNA binding proteins. The active sites for cleavage are NOT nearby the binding domain in the 3-D structure. Theses peptides can bind DNA on their own and their binding activity, like the native protein, is modulated by serine phosphorylation (Ser16 below).
Region 1 Region 2
< – – – – – – – – – – > < – – – – – – – – – – – – – – – – – – – – >
5 1 5 2 5 3 5
| | | |
1 Q H R R K N Q A R R S R L E R R V E I L V T E V H D L K K R L E G L
2 Q H K R R N Q A R R S R L A R K V E A L T A E V K A L R S E L N Q L
3 E H R R K N Q A R R S R L A Q K V S M L T A A V G T L R S E L D Q L
4 E H R R R N Q A R R S R L A I K V D S L T A A V M A L K A E L N R L
5 E H K R R N Q A R R S R L A I R V Q S L T A A V M T L K S E L N K L
6 E H K R R N E A R R S R L S V K V D A L V A E V M S L R S K L G Q L
7 Q H R R R N E A R R S R L Q E R V D N L S K E V R I L E K N L Q R L
8 Q H K R R N E A R R S R L Q Q R V E S L S N E V Q S L R D E L Q R L
9 Q H K R R N E A R R S R L G Q R V E A L K S E V S S L R I E L D R L
Indicate JUST the completely conserved residues in the spaces provided below
1)_ _ _ _ _ __ _ _ _ __ _ _ _ __ _ _ __ _ __ _ _ _ __ _ _ _ _ _
In (2) use the code, H=hydrophobic, P=polar-uncharged, += basic, – = acidic, to indicate the characteristics of ONLY the conserved residues (one per residue).
2)_ _ _ _ _ __ _ _ _ __ _ _ _ __ _ _ __ _ __ _ _ _ __ _ _ _ _ _
3) Based on your answers to (a) and (b) what secondary structure formed by Region 2 ?
Why do you think this?
4) Which region probably interacts with DNA? Why do you think this?
Specific labeling of proteins with 15N amino acids along with “NMR-editing” techniques allows one to observe only amide proton resonances for particular residues. The BDUN2 DNA binding domain (residues 5-39) was labeled with 15N amino acids in the four most N-terminal residues, Gln5-His6-Lys7-Arg8. The 15N-edited NOESY spectra (right) shows the differences in the main chain amide region cross peaks in the presence and absence of DNA. The phosphorylated sample looks the same as with no DNA. The N-H chemical shifts do change when DNA is added but this is not shown here. When the pH is varied from 5-9, the chemical shift for resonance 3 changes.
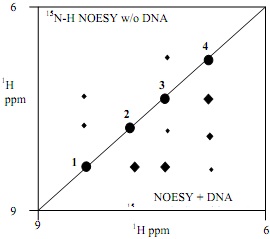
Without DNA, the greatest change occurs between 6 and 6.5, whereas with DNA present the greatest change occurs between pH 8 and 8.5.
5) Based on the NMR spectra, what protein structure is induced in Region 1 on DNA binding ?
6) Given your answer in (5), what topographic feature of the DNA double helix do these residues likely interact with? _
7) Which residue from the BDUN2 sequence is likely to give rise to resonance 3 (give type and number) ?
8) BRIEFLY (1-3 sentences) explain why this residue changes its titration behavior when DNA is added.
9) Calculate/Estimate the electrostatic contribution of this residue’s side chain (in the charged form) to the free energy of DNA binding ?Gbind?
Oligomerization, Function and stability.
Mature BDUN1 has 190 amino acids and by SDS PAGE has an apparent molecular weight of 20,000 Da. Using ultracentrifugation, native BDUN1 by has an apparent molecular weight of 40,000, but in the presence of 12 bp DNA forms a complex of MW~48,000. (Hint see cover page for important information-how much does the DNA weigh?). When it is phosphorylated on Ser16, the apparent molecular mass is 20,000. Free DNA is used in excess and is observed in both phosphorylated and un-phosphorylated experiments.
1) BRIEFLY (1-3 sentences) Describe and rationalize the oligomerization behavior of BDUN1, with and without DNA, and the effect of phosphorylation
Give two reasons why it is advantageous for BDUN1 bind DNA this way (from (1)) (IGNORE THE EFFECT OF PHOSPHORYLATION
4) Compared to free BDUN1, what happens to protein stability upon BDUN1 binding DNA? (increases/decreases)
5) What happens to protein stability upon BDUN1 phosphorylation? (increases/decreases)
6) What happens to DNA binding affinity upon BDUN1 phosphorylation? (becomes stronger/weaker)
7) The crystal structure of BDUN-1 with a 12 bp DNA site GAAGTGCACTCC (see below) shows no direct protein-DNA contacts with the central GpC base-step (underlined). Changing this to an TpA basestep, has little or no effect on DNA affinity. Circle the DNA structure below that most likely appears in the BDUN1/DNA crystal structure.
DNA binding analysis Using SELEX methods, BDUN1 through 5 bound the left group of aligned sequences while BDUN6 through 9 bound the right group. What are the “strong” consensus sequences?
BDUN 1-5 BDUN 6-9
AGATGCATCC GGACATGTCT
CGATCCATCG AGACTGGTCA
TGATTAATCT CGACCAGTCC
TGATATATCC TGACGGGTCG
GGATAAATCA GGACTCGTCA
Consensus 1)
3) What is significant about the two sequences ?
2)
4) By comparing sequences in the DNA-binding regions of each of the BDUN classes in (1), which residue, TYPE and NUMBER (e.g. “Glu27”) is likely responsible for the different DNA specificities?
5) A crystal structure of BDUN1 in complex with its consensus sequence (left above) shows the side chains from the residues given in (4)) interacting with two adjacent base pairs in the binding consensus sequences, (1) and (2). Given that the protein residue in BDUN6 is different, which base-steps in the above consensus sequences are likely contacted by each residue (5′?3′) (give only one step per consensus):
BDUN1 ____p____ BDUN6 ____p____
6) The crystal structure of BDUN6 was also solved with its consensus sequence (right, above) and with the exception of the differences in the DNA sequence, the structure appeared virtually identical in the vicinity of the “specificity-determining residue” you identified in (4).
Based on your knowledge of amino acid side chains and nucleotide base structures, sketch a hypothetical interaction of the two different amino acids from (4) interacting with the analogous base-steps you identified in (5), in such a way as to confer specificity for the corresponding binding sequences (draw just the bases and the amino acid with the relevant interactions).
PROTEIN FOLDING IN VIVO
Diagram the folding of a soluble protein in the cell starting from the ribosome. At each step, cells produce other proteins that increase the efficiency of folding. Answer the questions below.
1. In GENERAL, how do these proteins increase folding efficiency?
2. For each class of folding assistant, give their names, their specific substrates, and SPECIFICALLY how they increase folding efficiency of that step.
3. Some of these proteins are ATPases. Which ones and why do they have this activity?
4. Many of these proteins are overexpressed in heat-shocked cells. However, this also happens in the same way to 2% ethanol, cold shock, or even some mutated proteins. Explain why this diverse set of assaults leads to similar physiological outcomes?
