Describe the broad classification of prokaryotic and eukaryo
1. Indicate whether the following are referring to prokaryotic genomes or eukaryotic genomes or both.
a. Linear, Centriole, Nucleus, Chromosome, Nicked, Mitochondria, Mitosis or Meiosis, Scaffold, Histones, Circular, Supercoiled, Chloroplast, Plasmid, Chromatin, DNA dependent DNA polymerase, Asexual reproduction, Nucleoid, Ribosome, Binary fusion, Polycistronic, Poly-A tail signal sequence, Termination signal
2. Describe the broad classification of prokaryotic and eukaryotic genomes, indicating the various types of DNA molecules that are found in each and a detailed description of each of the DNA molecules.
3. Compare prokaryotic DNA with eukaryotic DNA, highlighting the differences between the two. What considerations do you need to take into account in a cloning experiment, to accommodate these differences?
4. Our research group is interested in identifying which wheat proteins are involved in aphid resistance responses. We want to develop an mRNA library to identify which genes are up-regulated during aphid infestation. Which lysis method would you recommend we use, justify your answer and explain the principle of the lysis method? Explain in detail how we would purify the mRNA from the lysate?
5. You take an optical density reading of a DNA sample but it is too concentrated to give you an accurate reading. You take 1 µL of the DNA sample and dilute it with 14 µL of distilled H2O. You then take aA260reading of the DNA on the spectrophotometer and it gives you an OD reading of 0.123 and a A280 reading which gives you an OD of 0.154. Written above the spectrophotometer is this conversion factor: 1 OD of dsDNA = 50 µg/mL of DNA.
a. What was the concentration of your original DNA sample?
b. Is this DNA sample suitable for PCR amplification? Explain your answer.
6. Agarose gel electrophoresis is a key tool in Molecular Biology. Describe the different applications you can use it for and what the key considerations are that you need to take into account for each application.
7. You need to prepare some solutions for a Southern blot. You are given 50X SSC, 10% SDS, and 5 M NaOH. How would you make 250 mL of a solution whose final concentrations were 1X SSC, 0.25% w/v SDS and 10 mMNaOH?
8. What factors need to be considered when designing a nucleic acid amplification experiment?
9. What are the major factors to consider when designing oligonucleotideprimers for PCR?
10. After amplifying the coding region of a gene of interest using RT-PCR, you run the amplified product on an agarose gel and find that no amplification occurred. What are possible factors that you need to consider during trouble shooting?
11. Compare PCR, RT-PCR and qPCR, highlighting the relevant template used, the components required, the principles of each and for which experiment you would use each of them.
12. What are the different types of restriction enzymes discovered to date and which type is normally used in Molecular Biology and why?
13. Name the different enzymes used in Molecular Biology and describe their functions.
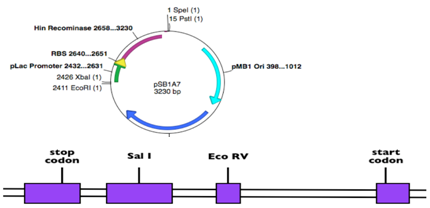
14. Below are a restriction map of a plasmid and a human gene of known sequence. Your task is to clone the human liver cDNA for this gene so that the start codon is closer to the EcoRI site than the PstI site of the polylinker/MCS, but you also need to get rid of the promoter+RBS+Hin insert already in the plasmid. Explain how you would accomplish the cloning using a series of numbered steps. Note that in the plasmid, the (1) refers to the frequency of these sites in the plasmid plus existing insert. In the gene’s restriction map, all the relevant restriction sites are indicated if they are present.
15. You have cloned a gene from the resurrection plant, Xerophytaviscosa. You wish to determine whether it is transcribed and translated under dehydration. Explain how you would do this.
16. You are a research scientist working on various aspects of firefly bioluminescence. In order to begin isolating the various genes involved in light production in this insect, it is necessary for you to establish a firefly gene library in E. coli. Taking into account the size of the firefly genome (approx. 1×105 kb), describe with the aid of a flow diagram how you would accomplish this task. Give reasons for the cloning strategy that you choose and the methodology that you follow.
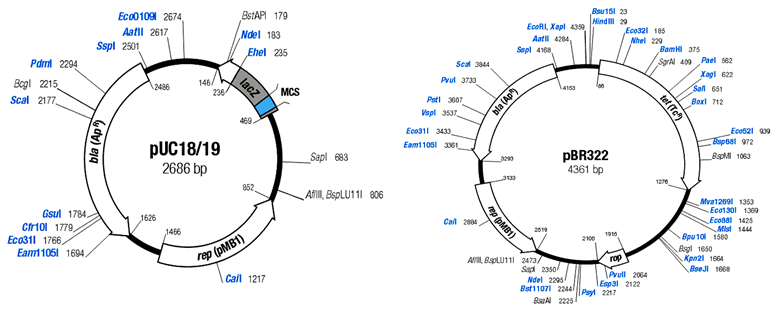
17. Describe the features of the plasmid cloning vector pUC19, the strategy one follows to obtain and identify E. colitransformants containing recombinant pUC19, and the advantages or disadvantages pUC19 has with respect to pBR322.
18. Write short notes describing the properties of cosmid cloning vectors, how they function, and the circumstances under which one would use such vectors when constructing gene libraries.
19. What is the difference between the methods used by the automated ABI sequencer and the Next Generation sequencers? Describe the principle of each method, its advantages and disadvantages.
20. You want to express a recombinant human protein in order to obtain sufficient quantities to perform kinetic experiments, obtain X-ray structure and confirm it role in a particular disease. Describe the steps you would follow in designing the cloning experiment/s, how you would confirm that you have the correct insert and that the protein is finally expressed. What additional considerations would you take if the protein is found to be toxic to bacteria and yeast cells.
21. Discuss the expression of a foreign gene, in bacteria also stating the problems encountered and how they can be overcomed.
22. The nucleotide sequence (5′ to 3′) for one strand of the double stranded coding region for Bradzyme gene is shown below. The coding region is underlined and the start (ATG) and stop codons (TGA) are in bold. The intervening sequence of 1590 bp is indicated between slahes (//). The pGLB expression vector you will be using to ligate the amplified insert into, contains a BamH1 (5’G^GATCC3′) and a Hind III (5’A^AGCTT3′) restriction site within the multiple clponing site.
23. 5′-gccaggctatggagcaggtgaatgagctttgt/1590 bp/agcaataaggccctgagtcggtgatattgatttga-3′
a. Design two oligonucleotide primers that could be used to amplify only the coding region, using the polymerase chain reaction (PCR), for insertion into the pGLB expression vector. Orientate each oligonucleotide 5′ to 3′. Indicate clearly what considerations were taken into account in the design of the primers.
b. Discuss what steps can be followed during problem solving of the PCR reaction when either (i) no bands or (ii) multiple bands are obtained after PCR reaction.
24. i) You have a strain of bacteria that requires amino acid lysine (lysine auxotroph). The strain is also sensitive to antibiotic streptomycin.
If you expose this strain to a mutagen
a. Explain how auxotrophic bacteria will be isolated
b. How would you isolate mutants that resist streptomycin?
ii) List the contents of a DNA sequencing reaction and describe what reactions occur within the sequencing tube and how the DNA sequence chromatogram is fully obtained
iii) Describe the possible errors/anomalities that may occur in sequence chromatograms and discuss what may have caused this problem